NeuralNav: Difference between revisions
No edit summary |
No edit summary |
||
| (6 intermediate revisions by one other user not shown) | |||
| Line 10: | Line 10: | ||
This project is funded under the NIH/NCI grant 1R01CA138419-01A1, as part of NCI academia-industry partnership initiative described in PAR-07-214, "Academic-Industrial Partnerships for Development and Validation of In Vivo Imaging Systems and Methods for Cancer Investigations." [http://grants.nih.gov/grants/guide/pa-files/PAR-07-214.html] | This project is funded under the NIH/NCI grant 1R01CA138419-01A1, as part of NCI academia-industry partnership initiative described in PAR-07-214, "Academic-Industrial Partnerships for Development and Validation of In Vivo Imaging Systems and Methods for Cancer Investigations." [http://grants.nih.gov/grants/guide/pa-files/PAR-07-214.html] | ||
== Software == | |||
This project is contributing most of its software development efforts to: | |||
* [http://www.slicer.org 3D Slicer] | |||
* [[TubeTK]] | |||
== Project notes == | |||
* [[NauralNavAmigoSetup|System setup in AMIGO]] | |||
= Investigators = | = Investigators = | ||
* Principle Investigators | * Principle Investigators | ||
** William "Sandy" Wells III, Ph.D., Harvard Medical School & Brigham and Women's Hospital | ** William "Sandy" Wells III, Ph.D., <em>Harvard Medical School & Brigham and Women's Hospital</em> | ||
** Stephen R. Aylward, Ph.D., Kitware | ** Stephen R. Aylward, Ph.D., <em>Kitware</em> | ||
* Neurosurgeons: | * Neurosurgeons: | ||
** Allan Friedman, M.D., Chief of Neurosurgery, Duke University | ** Allan Friedman, M.D., <em>Chief of Neurosurgery, Duke University</em> | ||
** Alexandra Golby, M.D., Neurosurgeon, Harvard Medical School & Brigham and Women's Hospital | ** Alexandra Golby, M.D., <em>Neurosurgeon, Harvard Medical School & Brigham and Women's Hospital</em> | ||
* Researchers: | * Researchers: | ||
** Matthew Toews, Ph.D., Harvard Medical School & Brigham and Women's Hospital | ** Matthew Toews, Ph.D., <em>Harvard Medical School & Brigham and Women's Hospital</em> | ||
* Program Officer: | * Program Officer: | ||
** Pushpa Tandon, Ph.D., National Cancer Institute, National Institutes of Health | ** Pushpa Tandon, Ph.D., <em>National Cancer Institute, National Institutes of Health</em> | ||
== Participating organizations == | == Participating organizations == | ||
| Line 34: | Line 44: | ||
| align="center" | [[image:NeuralNav-NCILogo.jpg|100px]] <br> NCI | | align="center" | [[image:NeuralNav-NCILogo.jpg|100px]] <br> NCI | ||
|} | |} | ||
== Related software == | == Related software == | ||
{| border="0" width="75%" cellpadding="20" align="center" | {| border="0" width="75%" cellpadding="20" align="center" | ||
| align="center" | [[image:NeuralNav-TubeTKLogo.jpg|100px]] [http://public.kitware.com/Wiki/TubeTK <br> TubeTK] | |||
| align="center" | [[image:NeuralNav-SlicerLogo.jpg|100px]] [http://www.slicer.org <br> Slicer] | | align="center" | [[image:NeuralNav-SlicerLogo.jpg|100px]] [http://www.slicer.org <br> Slicer] | ||
| align="center" | [[image:NeuralNav-IGSTKLogo.jpg|100px]] [http://www.igstk.org <br> IGSTK] | | align="center" | [[image:NeuralNav-IGSTKLogo.jpg|100px]] [http://www.igstk.org <br> IGSTK] | ||
| Line 56: | Line 55: | ||
| align="center" | [[image:NeuralNav-CMakeLogo.jpg|100px]] [http://www.cmake.org <br> CMake] | | align="center" | [[image:NeuralNav-CMakeLogo.jpg|100px]] [http://www.cmake.org <br> CMake] | ||
| align="center" | [[image:NeuralNav-CDashLogo.jpg|100px]] [http://www.cdash.org <br> CDash] | | align="center" | [[image:NeuralNav-CDashLogo.jpg|100px]] [http://www.cdash.org <br> CDash] | ||
| align="center" | [[image:NeuralNav-MIDASLogo.jpg|100px]] [http://www.insight-journal.org/midas <br> MIDAS] | | align="center" | [[image:NeuralNav-MIDASLogo.jpg|100px]] [http://www.insight-journal.org/midas <br> MIDAS] | ||
|| | || | ||
|- | |- | ||
|} | |} | ||
Latest revision as of 18:38, 5 December 2012
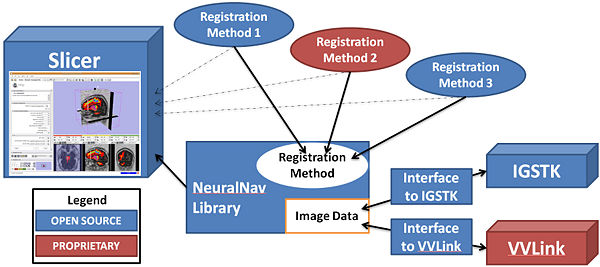
Neurosurgical navigation systems have reduced the risk of complications from surgery and have allowed surgeons to remove tumors that were once considered inoperable. However, many techniques used by neurosurgical navigation systems to align pre- and intra-operative images are inaccurate when tissue deformations occur. Deformations commonly arise from tumor resection, gravitational effects on the organ, and the use of hyperosmotic drugs. Deformable intra-operative image registration remains a significant challenge for neurosurgical guidance.
We proposed two new methods for registering pre-operative MRI with intra-operative 3D ultrasound data, during craniotomies for brain tumor resection. These methods will be delivered as part of an extensible NeuralNav toolkit that provides a common API for fetching tracker data and intra-operative images from commercial (VectorVision by BrainLAB) and research (Image-Guided Surgical Toolkit, IGSTK by Georgetown Univ) surgical guidance systems. Development and validation of methods and efforts will be conducted in collaboration with top neurosurgeons, the developers of IGSTK, and BrainLAB.
Funding
This project is funded under the NIH/NCI grant 1R01CA138419-01A1, as part of NCI academia-industry partnership initiative described in PAR-07-214, "Academic-Industrial Partnerships for Development and Validation of In Vivo Imaging Systems and Methods for Cancer Investigations." [1]
Software
This project is contributing most of its software development efforts to:
Project notes
Investigators
- Principle Investigators
- William "Sandy" Wells III, Ph.D., Harvard Medical School & Brigham and Women's Hospital
- Stephen R. Aylward, Ph.D., Kitware
- Neurosurgeons:
- Allan Friedman, M.D., Chief of Neurosurgery, Duke University
- Alexandra Golby, M.D., Neurosurgeon, Harvard Medical School & Brigham and Women's Hospital
- Researchers:
- Matthew Toews, Ph.D., Harvard Medical School & Brigham and Women's Hospital
- Program Officer:
- Pushpa Tandon, Ph.D., National Cancer Institute, National Institutes of Health
Participating organizations
Error creating thumbnail: Unable to save thumbnail to destination Brigham and Women's Hospital |
 Kitware |
 Harvard |
 Duke University |
BrainLAB |
 NCI |
Related software
 TubeTK |
 Slicer |
 IGSTK |
 CTK | |
Error creating thumbnail: Unable to save thumbnail to destination ITK |
Error creating thumbnail: Unable to save thumbnail to destination CMake |
 CDash |
MIDAS |